Renaissance Simulations Laboratory
Explore the first galaxies through physics-based modeling, simulation, and data exploration
User Guide
This user guide provides details on the various data products available for download on the RSL as well as performing analysis on the RSL itself.
Data Downloads
Snapshots, halo catalogs, and merger trees are available for download for
all of the simulations in the Rnaissance collection. A list of all simulations
is given on the Investigate page, with links to tables
of all existing data products. Halo catalogs have been created for every
snapshot and all are immediately available for download. However, due to their
large size (100-200 GB), only a subset of snapshots are currently hosted on the
RSL. Additional snapshots can be made available (for both download and analysis)
upon request, to the limit of available storage space on the RSL webserver.
Clicking the "request data" link next to a snapshot will open an email to the
RSL admins with the simulation and snapshot names. An account on the RSL is not
required to download data.
Snapshots
Snapshots contain all gas and particle data for the simulation at a single instant in time. A snapshot is stored on disk within a single directory, named something like RD00041, indicating that it’s the 42nd redshift-based output. In this directory is an ascii file containing all runtime parameters, a series of HDF5 files storing the grid and particle data, and some other files containing the layout of the adaptive mesh hierarchy and boundary conditions. These datasets can be loaded with yt by providing the path to the parameter file.
>>> import yt
>>> ds = yt.load("RD0041/RedshiftOutput0041")
Halo Catalogs
Halo catalogs for all snapshots have been created using the Rockstar-Galaxies halo finder. This creates two output formats, ascii and binary.
Ascii
Ascii-format halo catalogs are named out_#.list, where the number is
the n’th dataset run through Rockstar, not necessarily corresponding to
the snapshot with the same number. The file header contains a full
description of the data within.
Binary
Binary-format halo catalogs consist of a series of files named
halos_<snapshot name>.#.bin, there the snapshot name is the directory
name of that snapshot, and the number is the Rockstar processor number.
You will need to keep all of the *.bin files in order to load them.
This data can be
loaded with yt
by providing the path to the zero’th file.
>>> import yt
>>> ds = yt.load("halos_RD0041.0.bin")
Merger Trees
Merger trees have been created from the Rockstar halo catalogs using consistent-trees. Two formats are available for download, the raw consistent-trees output and ytree format. Both of these can be loaded with ytree.
consistent-trees
The consistent-trees file is named tree_0_0_0.dat and is an ascii
file. The file header contains a full description of the data and units.
This file can be loaded with ytree by doing:
import ytree
arbor = ytree.load("tree_0_0_0.dat")
ytree
The ytree format has identical contents to the consistent-trees
file, but is optimized for faster i/o. It consists of a directory
with a series of HDF5 files. The merger trees available on the RSL
have been named according to the simulation from which they were
created, but in all lowercase. They can be loaded by providing the
path to the HDF5 file of the same name:
import ytree
arbor = ytree.load("rarepeak_lwb/rarepeak_lwb.h5")
Analyzing on the RSL
The Renaissance Simulation Laboratory provides a Python environment with which to analyze all hosted data without needing to download anything. Computation is performed on the RSL webserver. This is facilitated by launching a Jupyter notebook server that has access to both the simulation data and persistent personal storage. Users can save notebooks and generated data products in their storage and download them to their local machine. The Python environment comes equipped with the yt and ytree analysis packages as well as all of their dependencies, including matplotlib, NumPy, and SciPy.
Account Creation and Login
An account is required for running analysis on the RSL. This account provides you with persistent storage for scripts and data. The first time you launch a Jupyter notebook session, you will be prompted to create a new account, either by providing a username and password or by authenticating with Globus or GitHub.
Launching Jupyter Notebook Sessions
Each simulation page contains two links for launching a Jupyter
notebook session. These are located in the top row of the upper
table, in the row labelled Analyze. Each of these are discussed
further below. If you are new to Jupyter notebooks, the documentation
can be found here.
Hit Enter in order to edit a notebook cell and shift+Enter to run
that cell.
Launch a Jupyter Session
The link entitled "Launch Jupyter" will start a regular Jupyter
notebook server. Clicking this will open a new tab with a file
browser showing two folders, data and home.
The data folder contains all data corresponding to the simulation
to be analyzed. The home folder is your storage space. Files stored
in home will be available from every Jupyter session you run. Click
on the folder names to navigate into them.
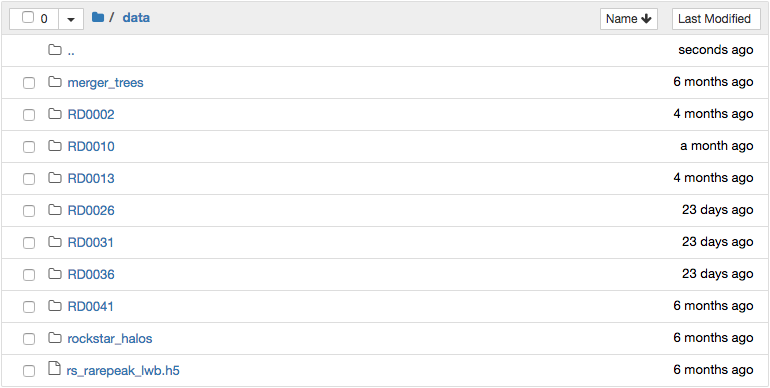
The data folder is read-only. Files can be download by clicking the
check box next to them and then clicking the Download button that
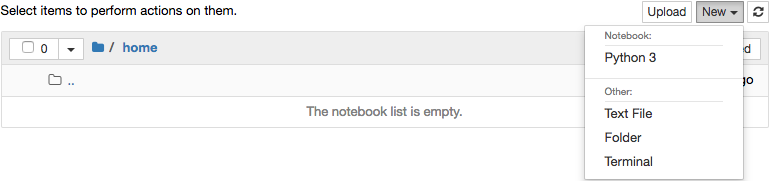
will appear above. New notebooks should be created from within the
home folder. Start a new notebook by going to the New menu in
the upper right and selecting Python 3 under the Notebook
heading. It is also possible to open a window with a conventional
terminal by selecting Terminal.
Make sure to save your notebook by selecting "Save and Checkpoint" in
the File menu or by clicking the
button. When you
finished, select "Close and Halt" from the File menu.
Launch a Tutorial Session
The link entitled "Launch Tutorial" will start a Jupyter notebook
server with the same setup as the "Launch Jupyter" link, except with
a tutorial notebook in your workspace.
Open the notebook by clicking on it.
The code in the welcome notebook can be re-run to reproduce the output
shown by hitting shift+Enter in each cell. The welcome notebook can
serve as a template for future analysis. After closing the notebook,
it can be moved to your home folder to be saved by clicking the check
box next to it and clicking the Move button that will appear above.
Help!
If you need any additional help, please contact the RSL admins.